Molecular Plant Testing
Our molecular plant testing services provide detailed, valuable insight into your operation, giving you the power to act. Early and rapid detection of plant pathogens provides advanced knowledge of their impact and spread. Confirming the variety of your plants or testing for pollen incompatibility gives you the tools you need to tailor your strategies – and make changes if needed. Protect your new sport or variety using the power of genomic analysis. These assurance measures help you make the best management decisions possible, protect your investment, and maximize your ROI.
Flexibility in Testing
No matter whether you want to test one tree or thousands, one virus or many, we have the technology and expertise to bring you affordable, personalized results. Our high throughput robotic equipment can run large batches of samples, or our low throughput equipment can test just a few samples at a time.
Rapid Results
High throughput sampling and robotic DNA or RNA extraction means we offer quick results and competitive pricing. Turnaround times are generally 2 weeks for most projects, and we’ll keep you updated every step of the way. Our in-house sequencing capabilities (available 2024) mean we can provide in-depth insights with a quick turn around time.
Customized, Expert Advice
Our Ph.D.-level professionals can help pinpoint the best sampling and testing strategy for your situation and quickly provide a customized quote for our services. We offer guidance on when and how to sample your plants for the best results, and simplify sampling with a sampling kit. Your data is analyzed by experts, and we provide a concise report at the conclusion of the project.

Qualterra’s state-of-the-art molecular plant testing technologies give you the tools to make the best management decisions possible.
Our Services
Virus Screening
Viruses, viroids, and phytoplasma cause more than $30 billion in damage annually around the world. These diseases can lie dormant (latent) in a tree for years, escaping detection, before environmental or biological conditions cause symptoms. However, by testing rootstock and scion mother plants before investing in a new planting, the risk of planting infected material can be greatly reduced. Similarly, testing symptomatic plants can confirm the cause to be a virus (compared to abiotic stress, for example), allowing you to manage the disease.
A collaborative approach. We work together with partners like CPCNW, WSU, and WSDA to bring effective testing to the industry. And as part of Qualterra’s continuing mission to innovate, Qualterra has been awarded Small Business and Innovation grants to compare several Little Cherry Disease (LCD) detection methods and develop high throughput methods for sensitive, quick turnaround and cost-effective detection.
Offered By Qualterra | ||||
ELISA | RT-PCR | RT-qPCR | RNAseq | |
Technology | Antigen | PCR | PCR | Sequencing |
Price | ✓ | ✓ | ✓✓ | ✓✓✓ |
Turnaround | ✓✓✓✓ | ✓✓✓✓ | ✓✓✓✓✓ | ✓ |
Number of Samples | ✓ | ✓✓ | ✓✓✓ | ✓✓✓ |
Sensivity | ✓ | ✓✓✓ | ✓✓✓✓ | ✓✓✓✓✓ |
Number of Diseases Detected per Test | ✓ | ✓ | ✓✓ | ✓✓✓✓✓ |
Confidence of Results | ✓ | ✓✓✓ | ✓✓✓✓ | ✓✓✓✓✓ |
Discovery Potential | X | X | X | ✓✓✓✓✓ |
There are three main virus screening technologies used in the industry: Antigen-based ELISA assays, PCR-based assays which includes RT-PCR and RT-qPCR, and sequencing-based approaches.
A list of common diseases we test for using RT-PCR, RT-qPCR, and sequencing can be found here: Qualterra Virus Screening Pathogens for Website 2023-10 (29kb)
A full list of all the plant pathogens we screen for can be found here: Qualterra Virus Database 2023 (60kb)
Virus Screening by RTqPCR

RT-PCR and RT-qPCR both provide a quick and accurate diagnosis of infected crops. These tests will identify only the viruses that are tested for, giving you sensitive detection without overcomplication. These methods are ideal for rapid screening of individual plants for up to four viruses at a time.
How it works: RT-PCR and RT-qPCR are both extremely sensitive tests which target a small section of genetic material unique to each virus tested. The target sequence is amplified through a process called polymerase chain reaction, until there is enough material to be accurately detected using fluorescent dyes. These two testing types are very similar; the difference between them is that RT-PCR provides a simple yes/no answer as to whether the virus is present in the sample, while RT-qPCR can provide a quantitative measure of how much virus is present.
For diagnosis of Little Cherry Virus-2 and X-Disease, we use protocols provided by the WSU Clean Plant Center Northwest. Every year, Qualterra participates in the WSU Testing Proficiency Program to be listed as a proficient testing site. Labs with current proficiency tests can provide validation of diseased blocks, allowing Washington State growers to participate in the tree removal cost sharing programs offered by WSU Extension and the WSDA. Visit the WSU Tree-Fruit Website for more information regarding management of Little Cherry Virus and X-Disease.
Here is an example of RT-qPCR Data:
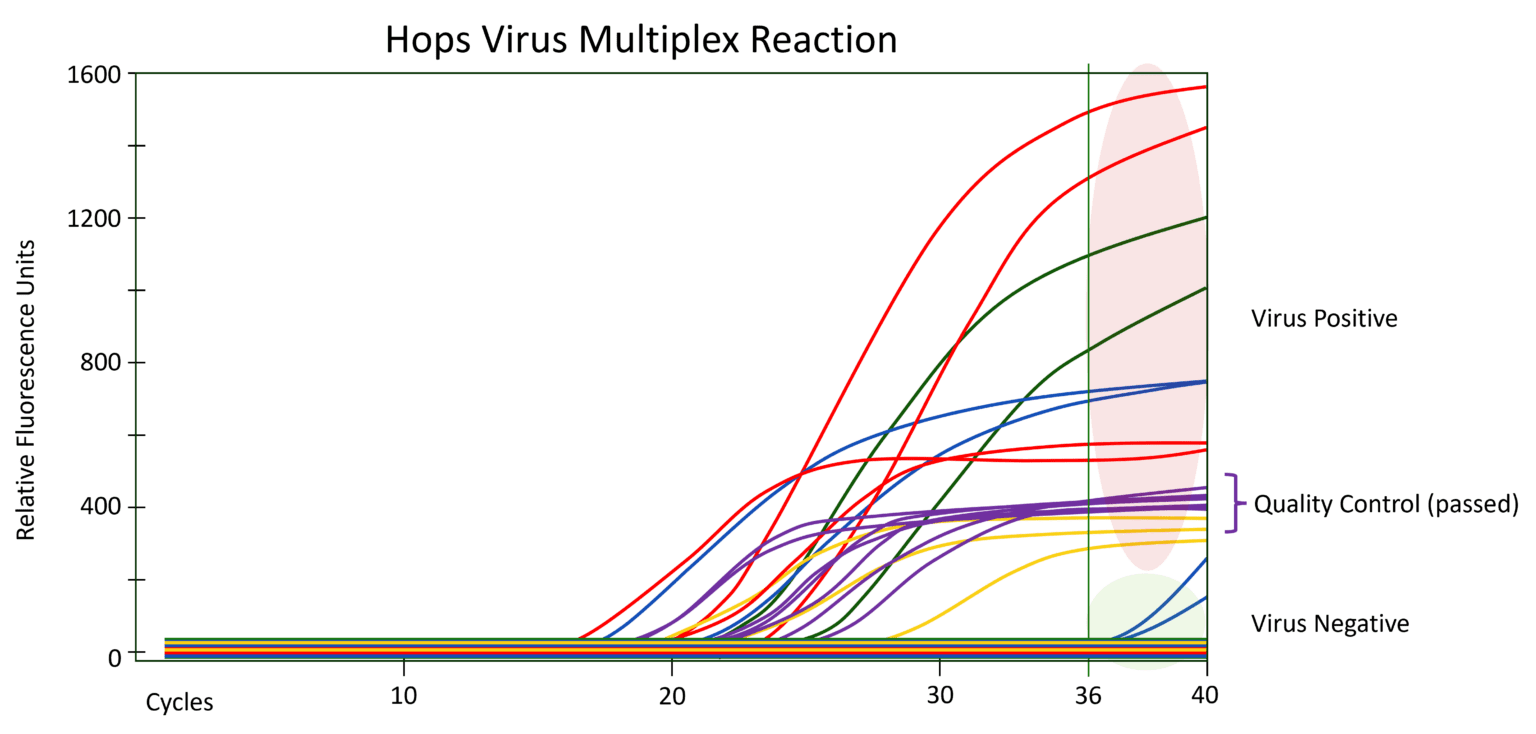
Fluorescent probe detection curves in RT-qPCR in Hops. Three viruses (Hop Latent Virus shown in red, American Hop Latent Virus shown in green, and Hop Mosaic Virus shown in blue) and one viroid (Hop Stunt Viroid shown in yellow) can be detected in a single reaction along with the RNA quality control marker NAD5 (shown in purple). In this case, curves that begin to amplify before cycle 36 are considered positive for the virus. Amplification curves that begin at earlier cycles indicate a higher titer of virus. No amplification curve indicates that the sample is negative for virus.
Virus Screening by Sequencing
Our most sensitive option for detection of viruses and viroids, RNA sequencing (RNAseq) combines enrichment for viruses with next-generation sequencing technology. All viruses and viroids present in the sample are detected, even at very low titers and before infected plants become symptomatic. This method is ideal for screening mother plants, new varieties, and pooled blocks of crops. Our technology is crop agnostic – we have extensive experience in horticultural crops, but any plant can be tested for viruses using this method.
How it works: Virus Screening by Sequencing starts with RNA from the plant tissue that is being screened, which contains both the plant’s native RNA and RNA from any other organism that was in the sample (including viruses or phytoplasma). We enrich viral RNA, and then the RNA is broken into smaller pieces. The sequencer then “reads” the sequence of each piece of RNA, producing a library of sequences. We can search the sample library against a database of known plant pathogenic virus sequences; if there are any hits, the sample is infected with that virus. We line these sequences up against the known genome of the virus, to visually confirm the infection.
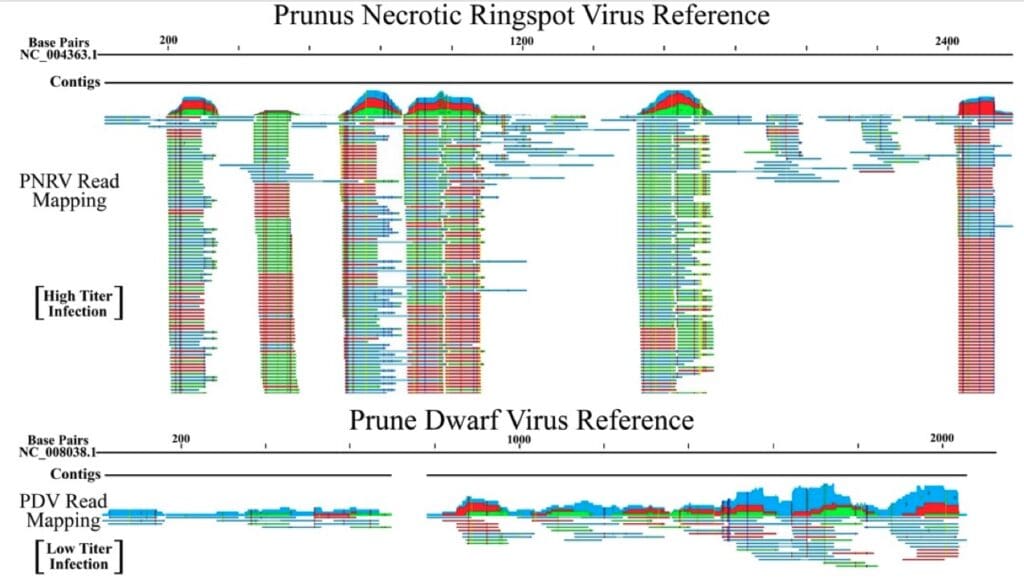
When a sample is infected with a virus, the library will contain sequences from that virus. Here, we show read mappings to a reference genome for two viral infections detected in sweet cherry: Prunus Necrotic Ringspot Virus (PNRSV, top) and Prune Dwarf Virus (PDV, bottom). The PNRSV infection was detected at high titer, indicated by a high read count and complete genome coverage (although it is normal for some areas to have more reads than other areas). PDV was detected with a relatively low read count, and incomplete genome coverage, indicating an active viral infection of low titer. When a sample is not infected with a particular virus, there will be no reads for that virus in the sample library.
RT-PCR | RT-qPCR | RNAseq | |
Price | ✓ | ✓✓ | ✓✓✓ |
Turnaround | ✓✓✓✓ | ✓✓✓✓✓ | ✓ |
Number of Samples | ✓✓ | ✓✓✓ | ✓✓✓ |
Sensitivity | ✓✓✓ | ✓✓✓✓ | ✓✓✓✓✓ |
Number of Diseases Detected per Test | ✓ | ✓✓ | ✓✓✓✓✓ |
Confidence of Results | ✓ | ✓✓✓ | ✓✓✓✓✓ |
Discovery Potential | ✓✓✓✓✓ |
True to type / Variety Verification testing
Mixups do happen … we’re all human. Our True-To-Type testing services deliver fast, reliable results to help maintain accuracy in the nursery or field. Our offerings are commonly used to check blocks for trueness to type, determine the identity of an unknown variety, or establish a fingerprint and protect a new variety.
How it works: We use Single Nucleotide Polymorphism (SNP) genotyping to verify plant variety; SNPs are single-base changes to the genome which can be detected by targeted PCR reactions. By looking at several SNPs, we can generate a “fingerprint” for each variety, and compare fingerprints to determine whether two plants are the same variety. Qualterra has an extensive database of fingerprints for Apple, Cherry, Grape, Hops, and Pear, and we source new varieties annually to update and validate our database.
(a)
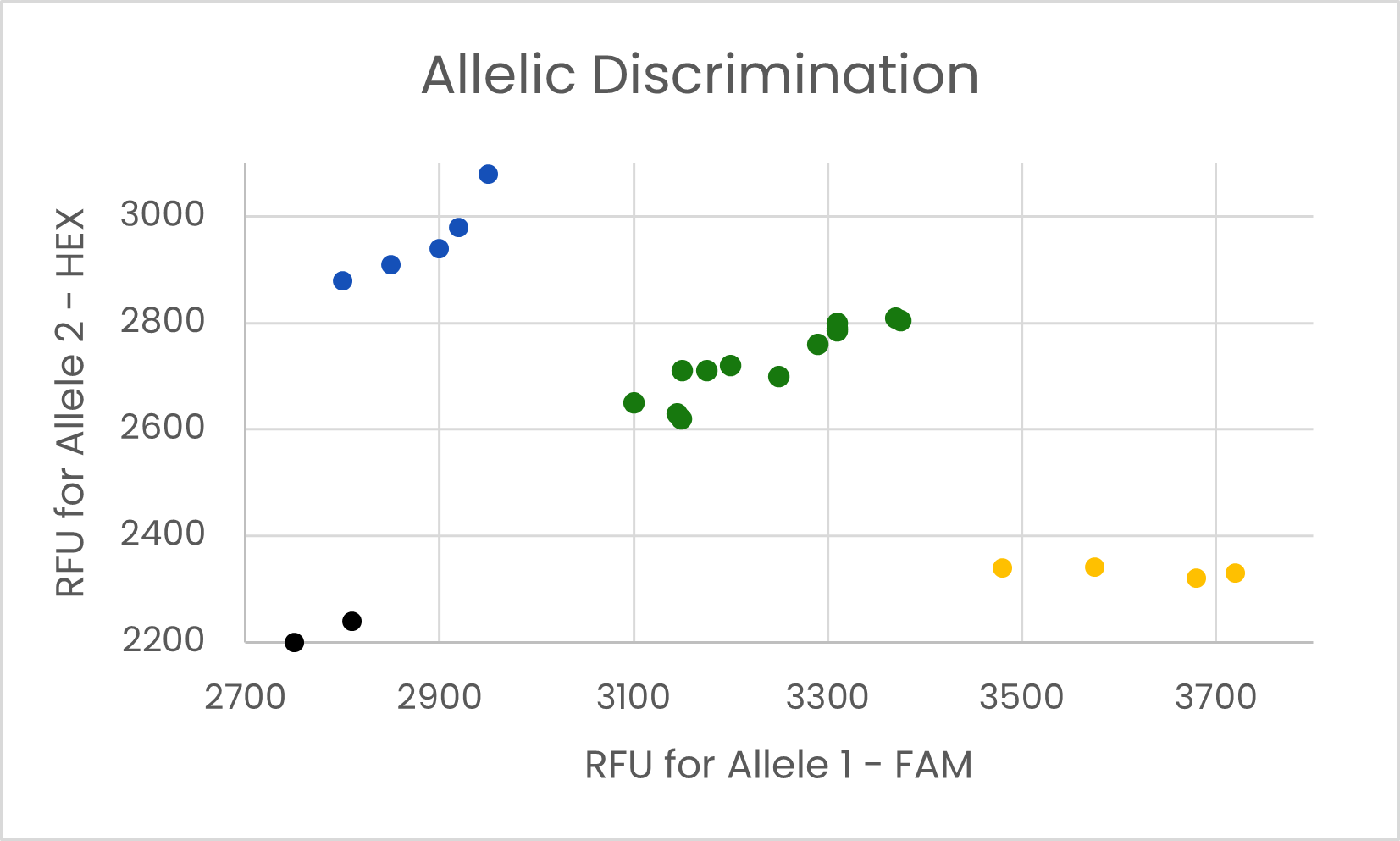
(b)
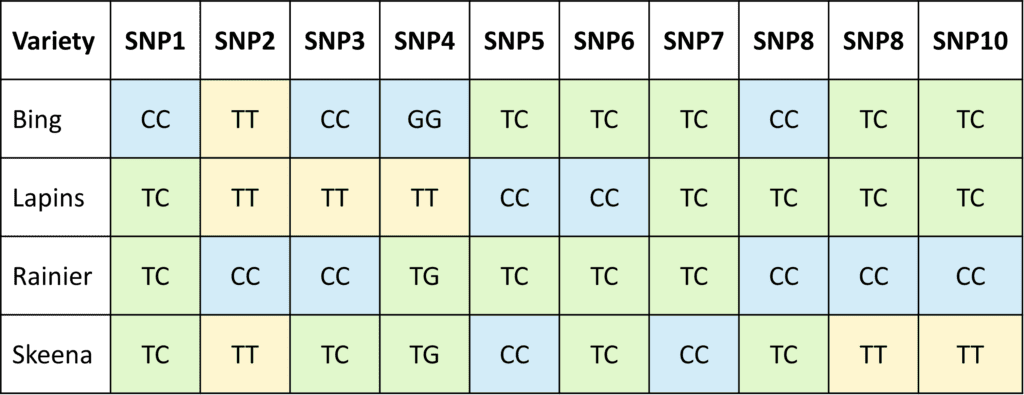
We use 7-20 SNP Markers to generate a fingerprint for each variety. (a) Each SNP is detected by qPCR, and (b) the results for all SNPs are compiled to generate a fingerprint for each variety. We can determine if an unknown sample matches one of the known fingerprints in the database, or if it is unique.
S-Allele Testing

Flowering plants employ several mechanisms to prevent self-fertilization and encourage outcrossing. Self-Incompatibility is one strategy, which prevents fertilization of a flower if the pollen has the same genetic basis as the stigma. By testing to determine which version (allele) of this gene the variety contains, one can determine which varieties can provide compatible pollen, and which varieties would be genetically incompatible. This testing can also identify varieties that contain alleles conferring self-fertility, such as the S4’ allele in cherry.
Variety | Allele Composition | Incompatibility Group |
Bing | S3S4 | III |
Compact Stella | S3S4' | Self-compatible |
Vista | S2S5 | VIII |
Lapins | S1S4' | Self-compatible |
Other Services

Sport / New Variety Verification.
While most breeding lines can be distinguished from their parents using traditional marker analysis (such as SNP marker analysis), a new sport mutant of an existing variety requires a different strategy. We use whole genome sequencing to identify small changes between the genome of the sport and its mother. This information can be used to show the novelty of the sport; additionally, we can design and validate assays allowing easy SNP testing in the future.
DNA / RNA Robotic Extraction services.
Our robotic nucleic acid extraction robot uses magnetic bead technology to effectively purify DNA or RNA from many plant tissues, including leaf, bark, or stem. Samples are prepared in 96-well plates, up to 8 plates at a time. Nucleic acids purified from this method can be used for a variety of applications, including PCR/RT-PCR/qPCR/RT-qPCR or Next Generation Sequencing.
Capillary Gel Electrophoresis services.
Our high-resolution capillary gel electrophoresis instrument can analyze up to 96 samples per run, with no dilution required. Gather accurate data, generate high-quality figures, or perform NGS sample quality control.
DNA Sequencing Services.
We work with sequencing service centers and leverage our own in-house sequencer (available 2024) to bring a wide array of DNA and RNA sequencing services to the industry. Common uses include protecting a new variety, molecular breeding, and marker development.


